Análisis de regresión lineal múltiple
Gonzalo Pérez
31 de marzo de 2020
3.
El objetivo de los siguientes datos es analizar si el consumo de ciertos alimentos y otros factores (como edad, sexo, etc.) tienen una relación con el nivel de plasma beta-carotene. El interés original de estos datos fue recabar información que pudiera servir para encontrar los factores asociados con niveles bajos de plasma beta-carotene dado que estos podrían estar asociados con el riesgo de desarrollar algunos canceres.
Variables:
- Dependiente. BETAPLASMA, Plasma beta-carotene (ng/ml)
- Explicativas:
- AGE (years)
- SEX (1=Male, 2=Female)
- VITUSE, Vitamin Use (1=Yes, fairly often, 2=Yes, not often, 3=No)
- CALORIES, Number of calories consumed per day.
- FAT, Grams of fat consumed per day.
- FIBER, Grams of fiber consumed per day.
- ALCOHOL, Number of alcoholic drinks consumed per week.
- CHOLESTEROL, Cholesterol consumed (mg per day).
- BETADIET, Dietary beta-carotene consumed (mcg per day).
Datos=read.table("images/archivos/prdata.dat",
header=FALSE, sep="\t")
names(Datos)=c("AGE", "SEX", "SMOKSTAT", "QUETELET", "VITUSE", "CALORIES", "FAT", "FIBER", "ALCOHOL", "CHOLESTEROL", "BETADIET", "RETDIET", "BETAPLASMA", "RETPLASMA")
head(Datos)str(Datos)## 'data.frame': 315 obs. of 14 variables:
## $ AGE : int 64 76 38 40 72 40 65 58 35 55 ...
## $ SEX : int 2 2 2 2 2 2 2 2 2 2 ...
## $ SMOKSTAT : int 2 1 2 2 1 2 1 1 1 2 ...
## $ QUETELET : num 21.5 23.9 20 25.1 21 ...
## $ VITUSE : int 1 1 2 3 1 3 2 1 3 3 ...
## $ CALORIES : num 1299 1032 2372 2450 1952 ...
## $ FAT : num 57 50.1 83.6 97.5 82.6 56 52 63.4 57.8 39.6 ...
## $ FIBER : num 6.3 15.8 19.1 26.5 16.2 9.6 28.7 10.9 20.3 15.5 ...
## $ ALCOHOL : num 0 0 14.1 0.5 0 1.3 0 0 0.6 0 ...
## $ CHOLESTEROL: num 170.3 75.8 257.9 332.6 170.8 ...
## $ BETADIET : int 1945 2653 6321 1061 2863 1729 5371 823 2895 3307 ...
## $ RETDIET : int 890 451 660 864 1209 1439 802 2571 944 493 ...
## $ BETAPLASMA : int 200 124 328 153 92 148 258 64 218 81 ...
## $ RETPLASMA : int 915 727 721 615 799 654 834 825 517 562 ...Usaremos, por simplicidad, variables binarias.
Datos$SEXFem=1*(Datos$SEX==2)
Datos$VITUSEYes=1*(Datos$VITUSE==1)
Datos$SEXFem=as.factor(Datos$SEXFem)
Datos$VITUSEYes=as.factor(Datos$VITUSEYes)
DatosRed=Datos[,c("BETAPLASMA", "AGE", "SEXFem", "VITUSEYes", "CALORIES", "FAT", "FIBER", "ALCOHOL", "CHOLESTEROL", "BETADIET")]
summary(DatosRed)## BETAPLASMA AGE SEXFem VITUSEYes CALORIES FAT
## Min. : 0 Min. :19 0: 42 0:193 Min. : 445 Min. : 14
## 1st Qu.: 90 1st Qu.:39 1:273 1:122 1st Qu.:1338 1st Qu.: 54
## Median : 140 Median :48 Median :1667 Median : 73
## Mean : 190 Mean :50 Mean :1797 Mean : 77
## 3rd Qu.: 230 3rd Qu.:62 3rd Qu.:2100 3rd Qu.: 95
## Max. :1415 Max. :83 Max. :6662 Max. :236
## FIBER ALCOHOL CHOLESTEROL BETADIET
## Min. : 3 Min. : 0 Min. : 38 Min. : 214
## 1st Qu.: 9 1st Qu.: 0 1st Qu.:155 1st Qu.:1116
## Median :12 Median : 0 Median :206 Median :1802
## Mean :13 Mean : 3 Mean :242 Mean :2186
## 3rd Qu.:16 3rd Qu.: 3 3rd Qu.:309 3rd Qu.:2836
## Max. :37 Max. :203 Max. :901 Max. :9642library(GGally)
ggpairs(DatosRed[,c(1,3,4)])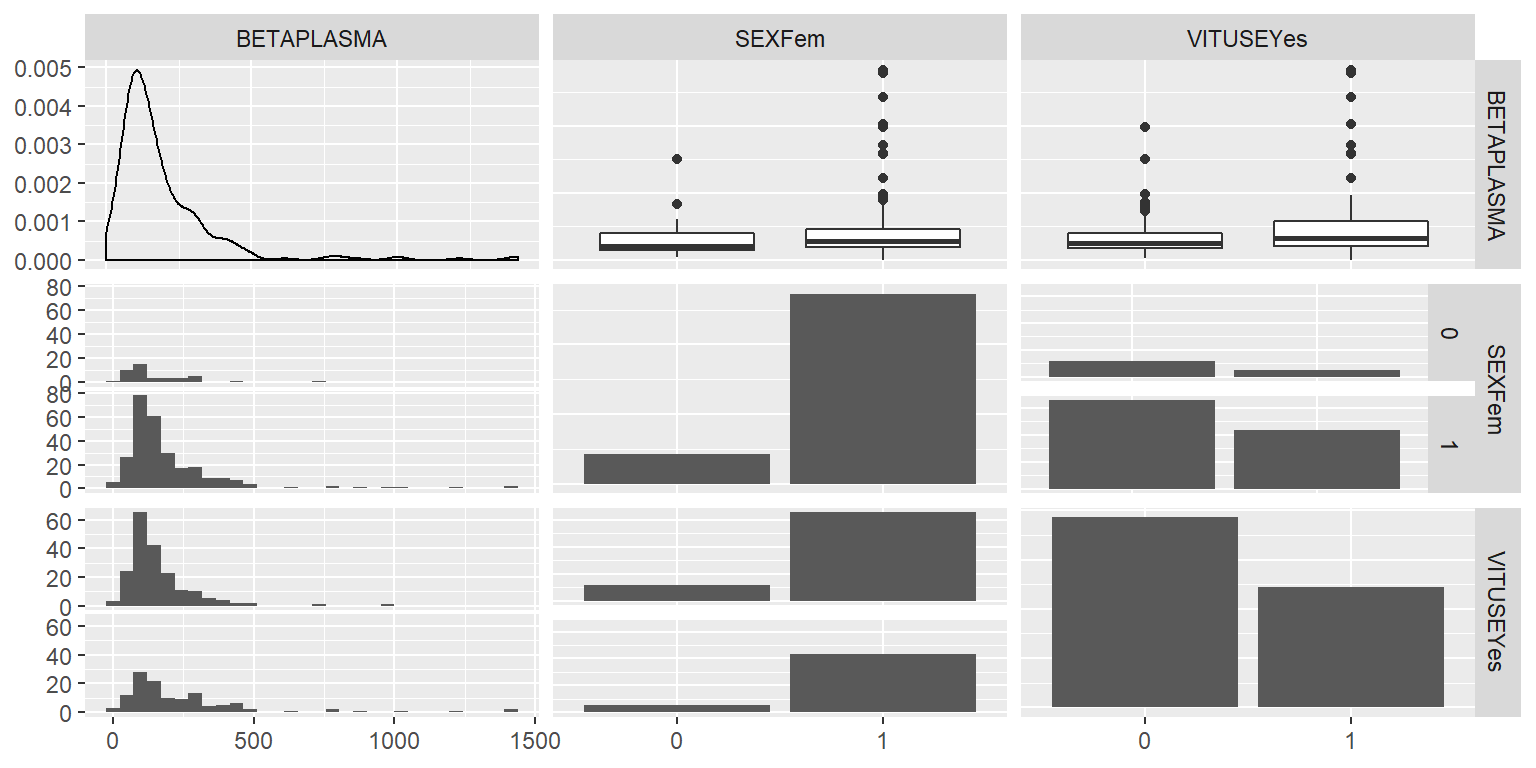
ggpairs(DatosRed[,c(1,2,5:10)])
DatosRed[ DatosRed$ALCOHOL>100, ]DatosRed[ DatosRed$CALORIES>4000, ]DatosRed[ DatosRed$BETAPLASMA==0, ]DatosRed2=DatosRed[-c(62, 257), ]library(GGally)
ggpairs(DatosRed2[,c(1,3,4)])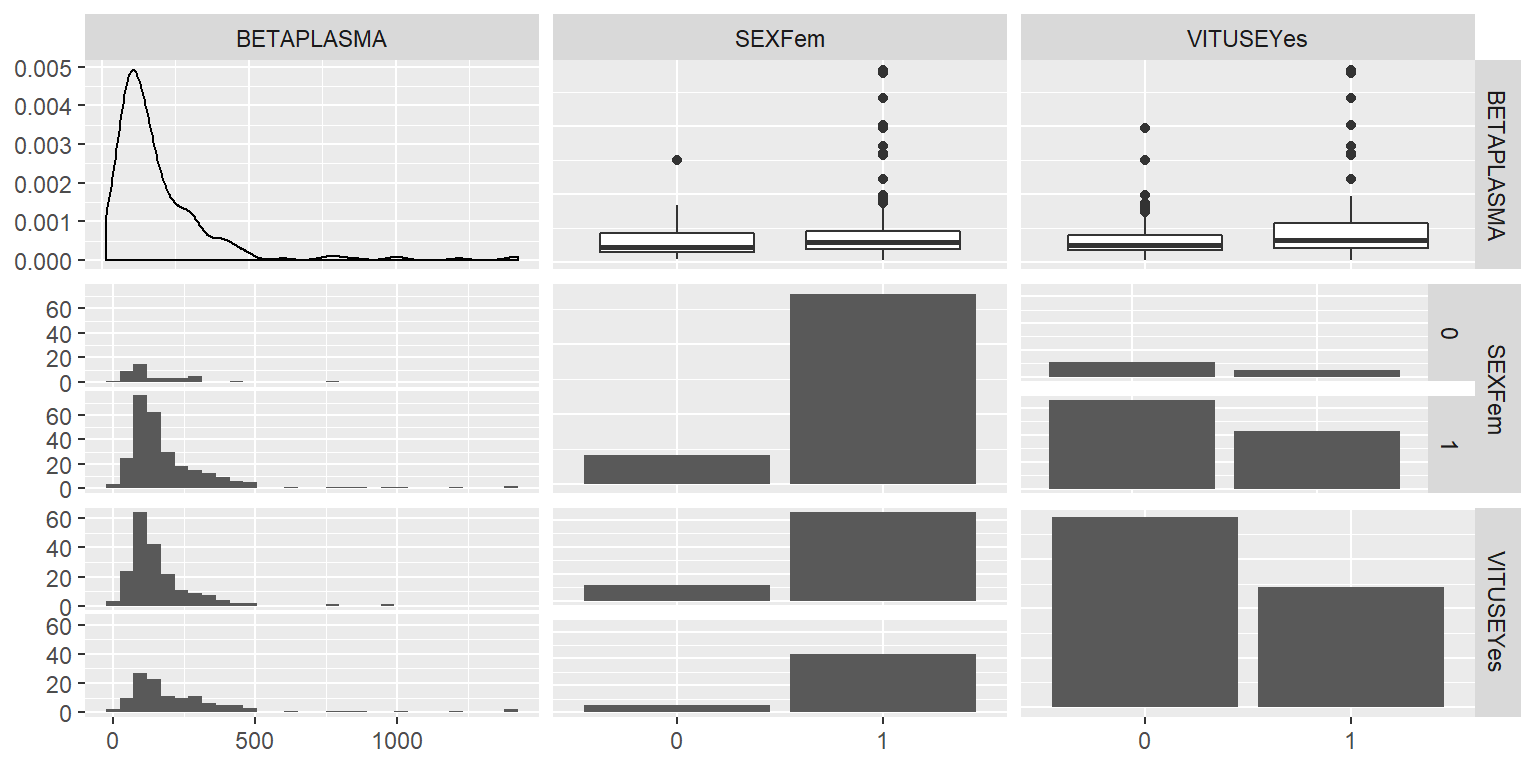
ggpairs(DatosRed2[,c(1,2,5:10)])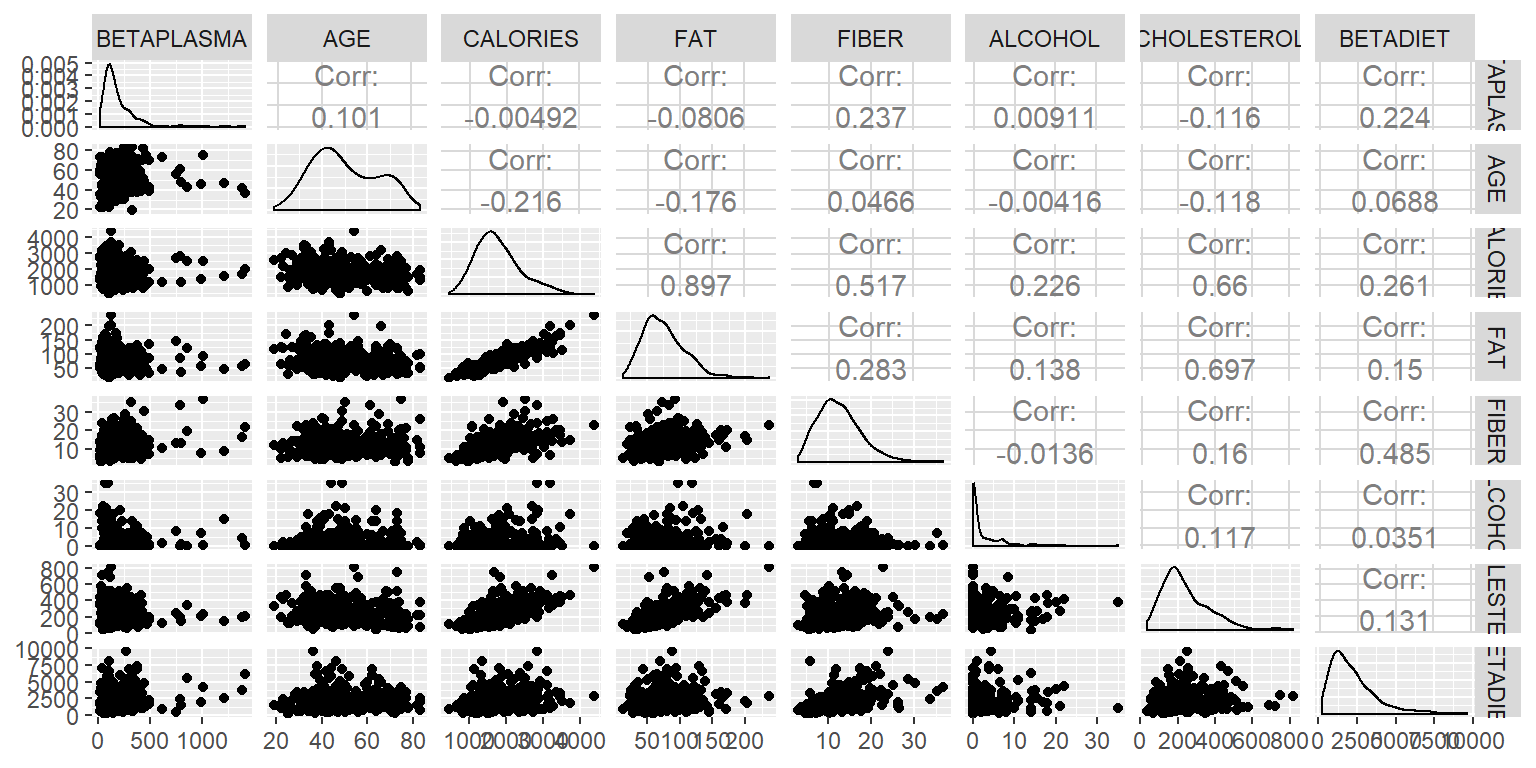
fit1=lm(BETAPLASMA~AGE+SEXFem+VITUSEYes+CALORIES+FAT+FIBER+ALCOHOL+CHOLESTEROL+BETADIET, data=DatosRed2)
summary(fit1)##
## Call:
## lm(formula = BETAPLASMA ~ AGE + SEXFem + VITUSEYes + CALORIES +
## FAT + FIBER + ALCOHOL + CHOLESTEROL + BETADIET, data = DatosRed2)
##
## Residuals:
## Min 1Q Median 3Q Max
## -295.3 -87.6 -35.1 37.1 1090.5
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 21.09796 68.90407 0.31 0.75967
## AGE 0.74956 0.75417 0.99 0.32107
## SEXFem1 47.95033 32.38050 1.48 0.13969
## VITUSEYes1 71.25656 20.50979 3.47 0.00059 ***
## CALORIES -0.02716 0.05176 -0.52 0.60013
## FAT -0.04179 0.81593 -0.05 0.95918
## FIBER 7.68289 2.84322 2.70 0.00728 **
## ALCOHOL 3.09462 2.21167 1.40 0.16277
## CHOLESTEROL -0.10536 0.11289 -0.93 0.35144
## BETADIET 0.01514 0.00761 1.99 0.04747 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 172 on 303 degrees of freedom
## Multiple R-squared: 0.147, Adjusted R-squared: 0.122
## F-statistic: 5.81 on 9 and 303 DF, p-value: 1.79e-07confint(fit1)## 2.5 % 97.5 %
## (Intercept) -1.1e+02 156.689
## AGE -7.3e-01 2.234
## SEXFem1 -1.6e+01 111.669
## VITUSEYes1 3.1e+01 111.616
## CALORIES -1.3e-01 0.075
## FAT -1.6e+00 1.564
## FIBER 2.1e+00 13.278
## ALCOHOL -1.3e+00 7.447
## CHOLESTEROL -3.3e-01 0.117
## BETADIET 1.7e-04 0.030plot(fit1)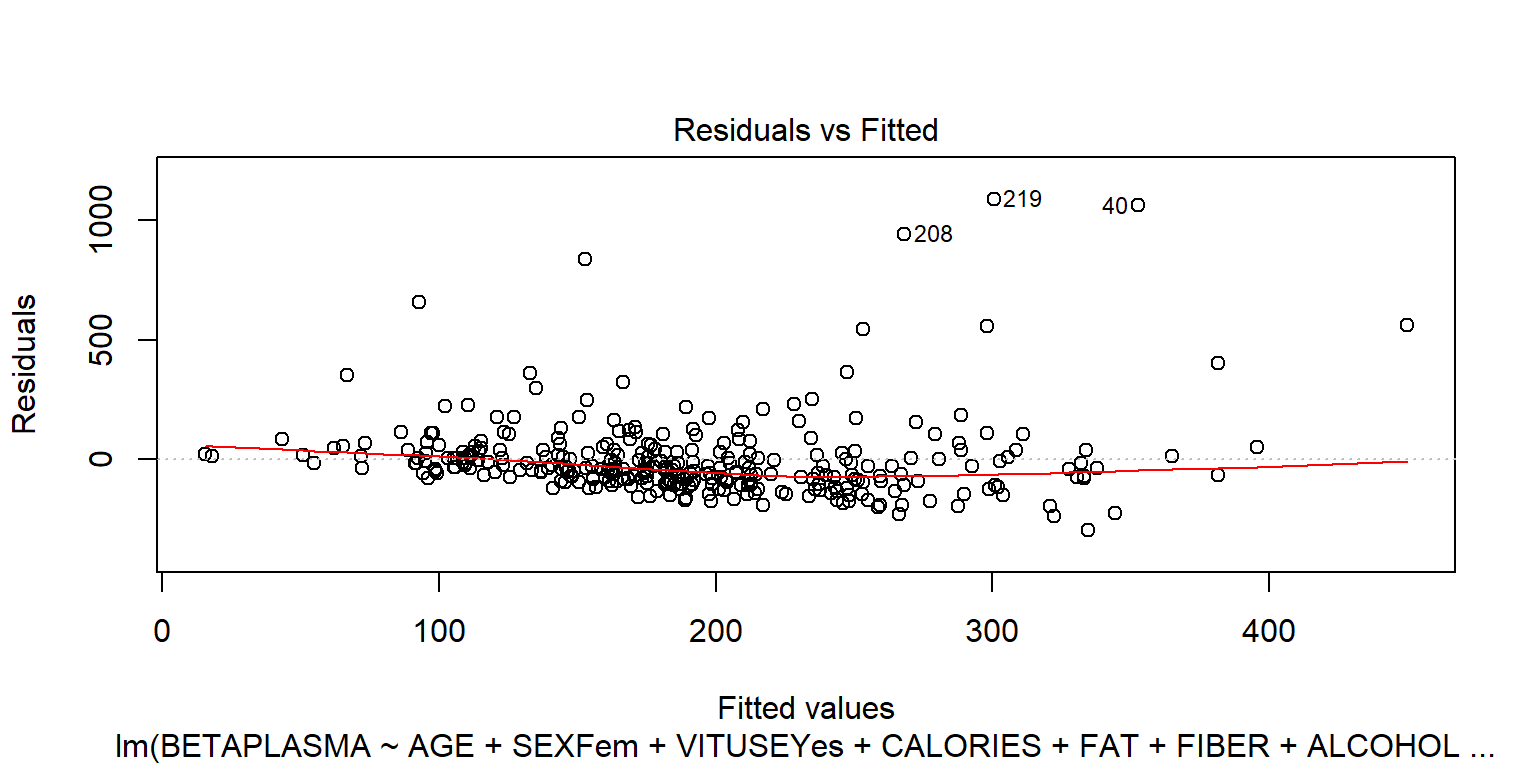


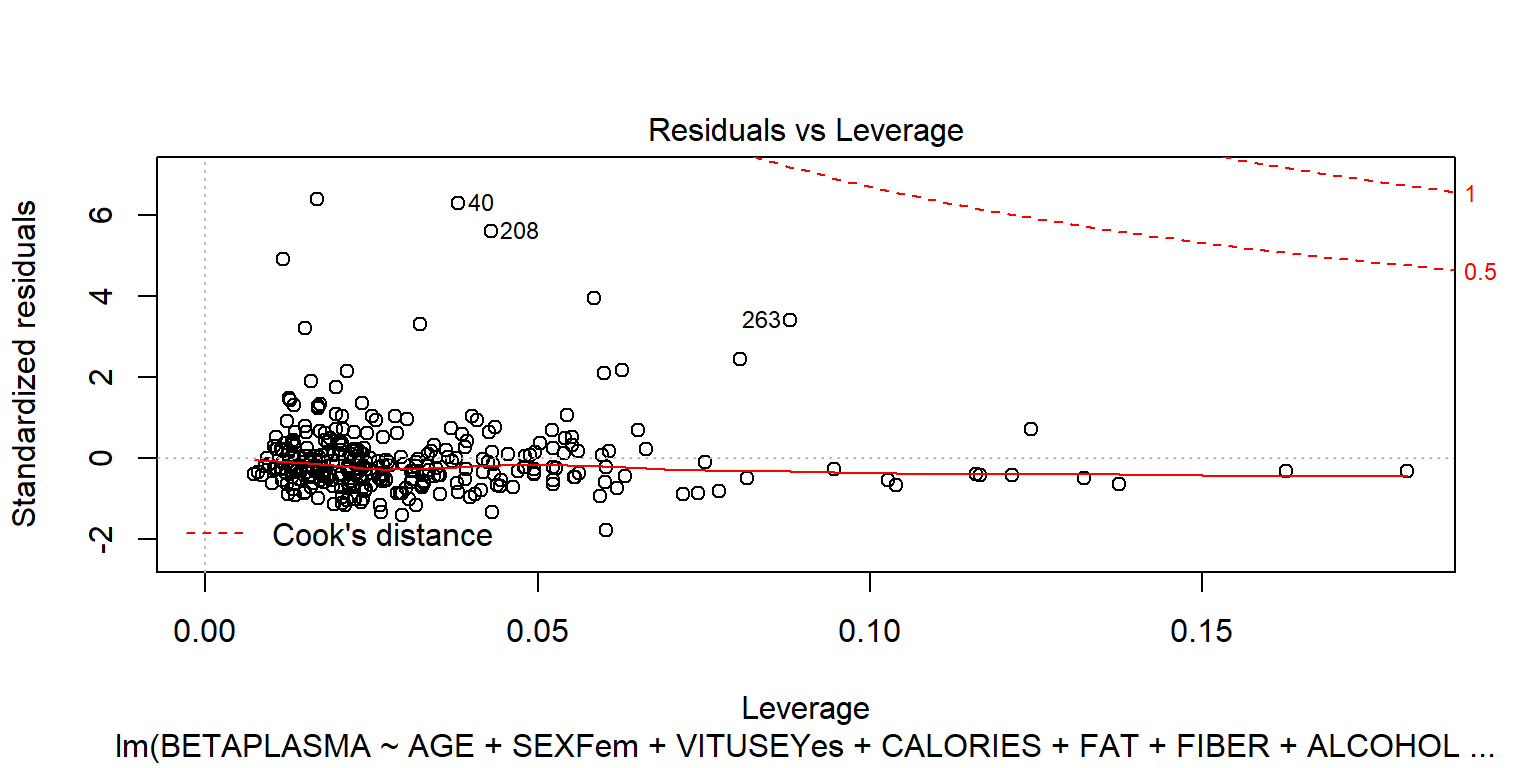
plot(fit1, 4)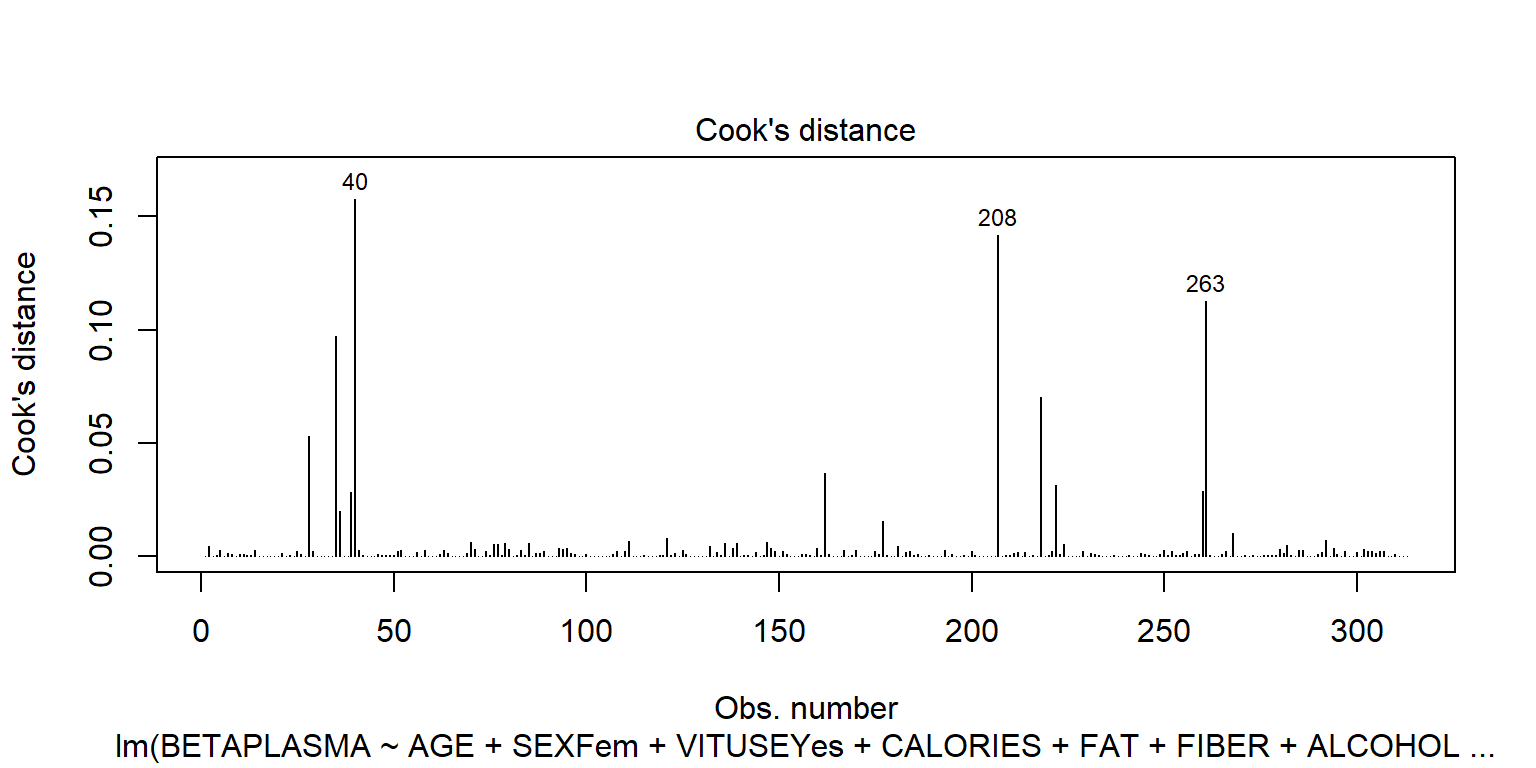
library(car)
fit1Ymod=boxCox(fit1, plotit=TRUE)
DatosRed2$BETAPLASMAlog=log(DatosRed2$BETAPLASMA+10)
fit2=lm(BETAPLASMAlog~AGE+SEXFem+VITUSEYes+CALORIES+FAT+FIBER+ALCOHOL+CHOLESTEROL+BETADIET, data=DatosRed2)
summary(fit2)##
## Call:
## lm(formula = BETAPLASMAlog ~ AGE + SEXFem + VITUSEYes + CALORIES +
## FAT + FIBER + ALCOHOL + CHOLESTEROL + BETADIET, data = DatosRed2)
##
## Residuals:
## Min 1Q Median 3Q Max
## -1.8298 -0.3546 -0.0217 0.3707 2.0218
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 4.27e+00 2.56e-01 16.69 <2e-16 ***
## AGE 5.05e-03 2.80e-03 1.81 0.0721 .
## SEXFem1 2.80e-01 1.20e-01 2.33 0.0206 *
## VITUSEYes1 2.24e-01 7.61e-02 2.94 0.0035 **
## CALORIES -1.69e-04 1.92e-04 -0.88 0.3799
## FAT 4.61e-04 3.03e-03 0.15 0.8792
## FIBER 3.49e-02 1.06e-02 3.30 0.0011 **
## ALCOHOL 8.05e-03 8.21e-03 0.98 0.3277
## CHOLESTEROL -4.18e-04 4.19e-04 -1.00 0.3193
## BETADIET 4.19e-05 2.83e-05 1.48 0.1386
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.64 on 303 degrees of freedom
## Multiple R-squared: 0.164, Adjusted R-squared: 0.14
## F-statistic: 6.62 on 9 and 303 DF, p-value: 1.21e-08confint(fit2)## 2.5 % 97.5 %
## (Intercept) 3.8e+00 4.8e+00
## AGE -4.6e-04 1.1e-02
## SEXFem1 4.3e-02 5.2e-01
## VITUSEYes1 7.4e-02 3.7e-01
## CALORIES -5.5e-04 2.1e-04
## FAT -5.5e-03 6.4e-03
## FIBER 1.4e-02 5.6e-02
## ALCOHOL -8.1e-03 2.4e-02
## CHOLESTEROL -1.2e-03 4.1e-04
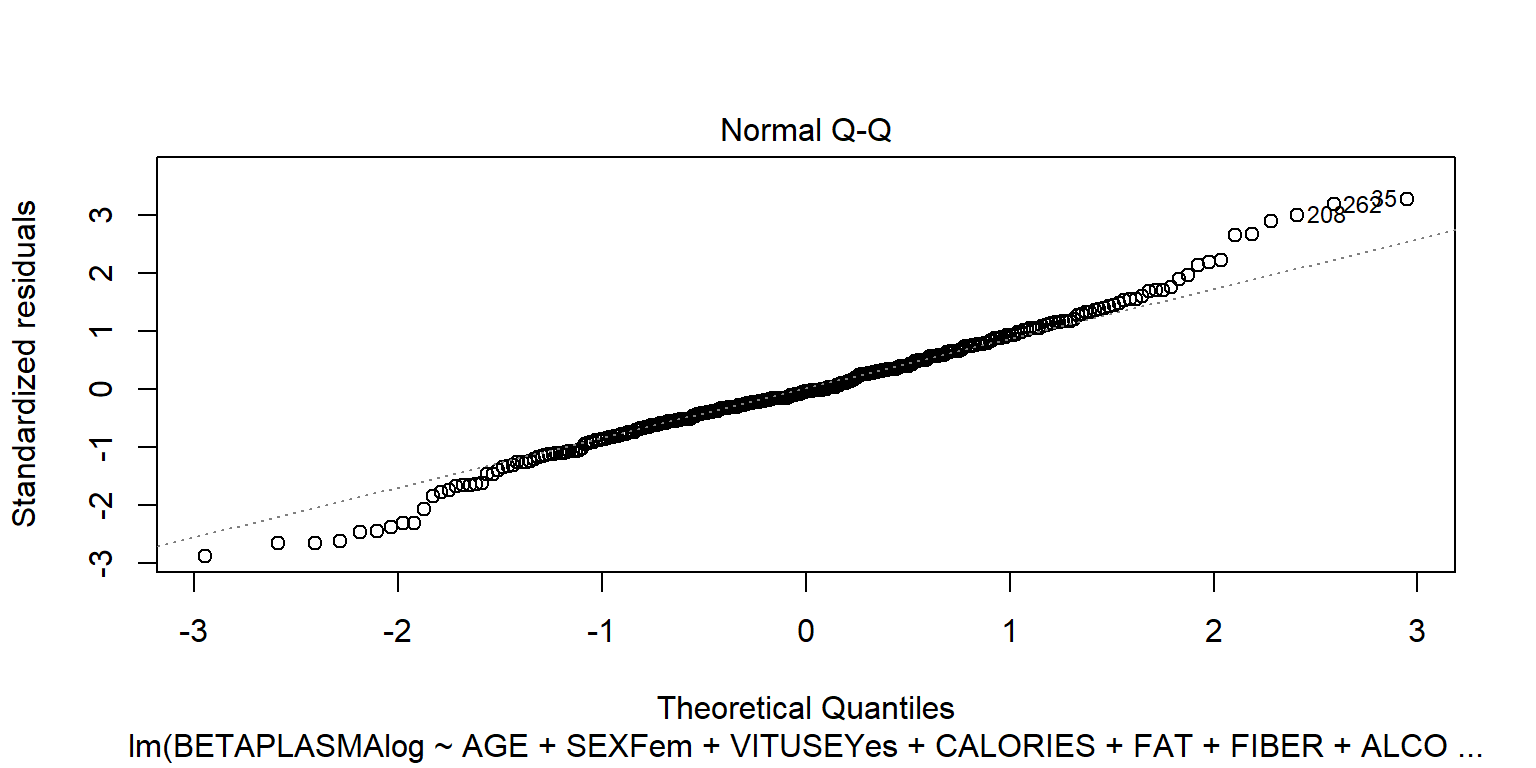
## BETADIET -1.4e-05 9.8e-05plot(fit2)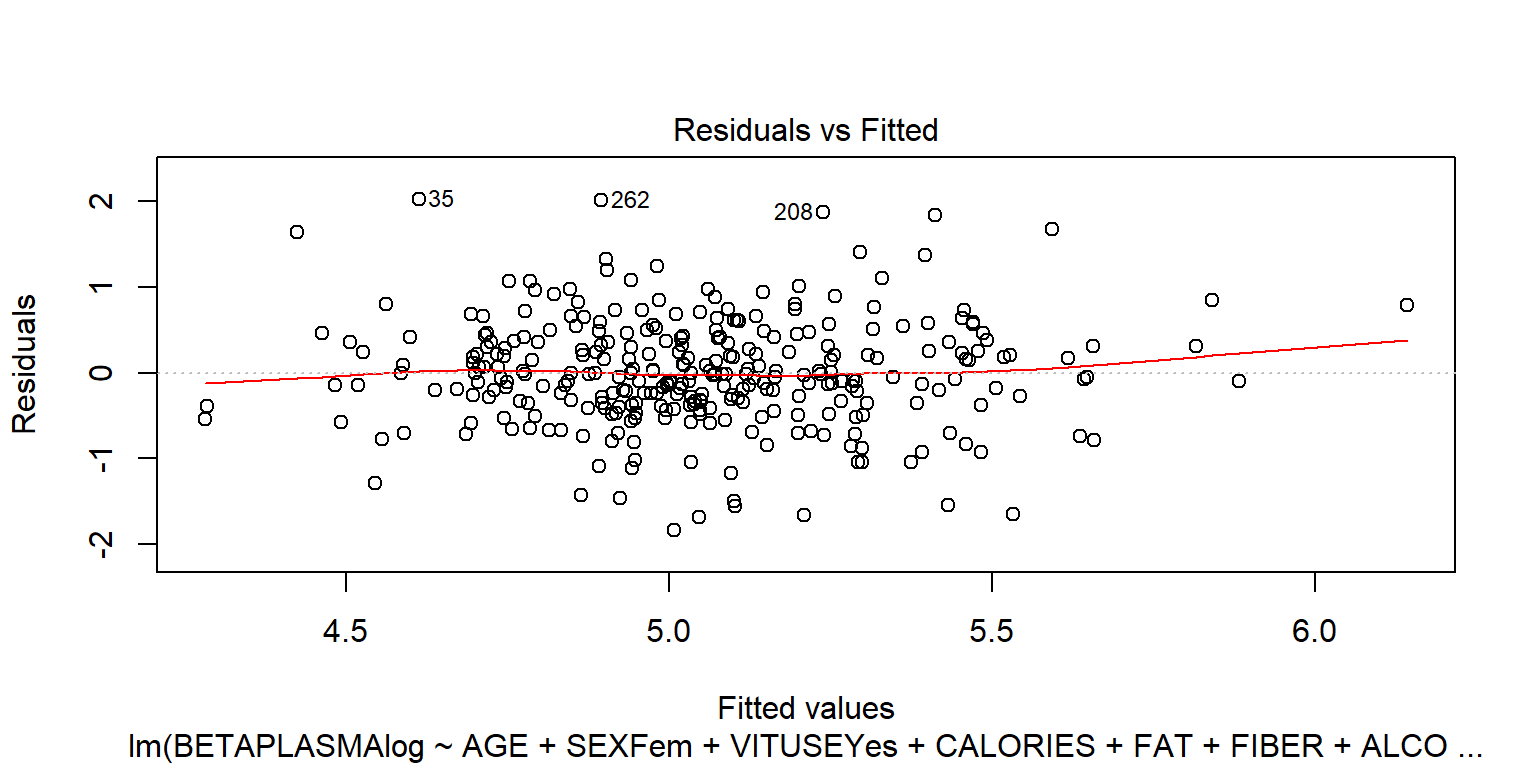
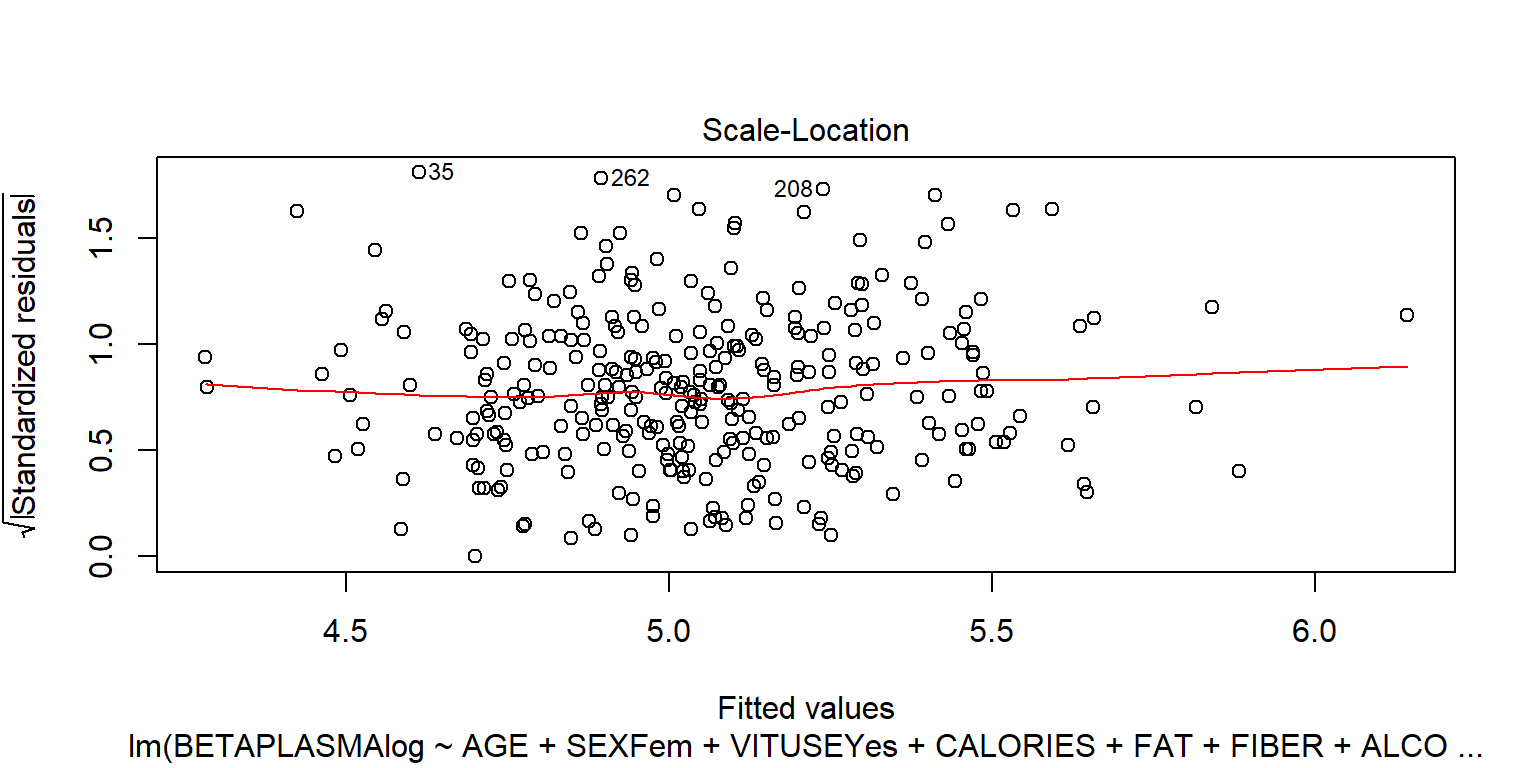
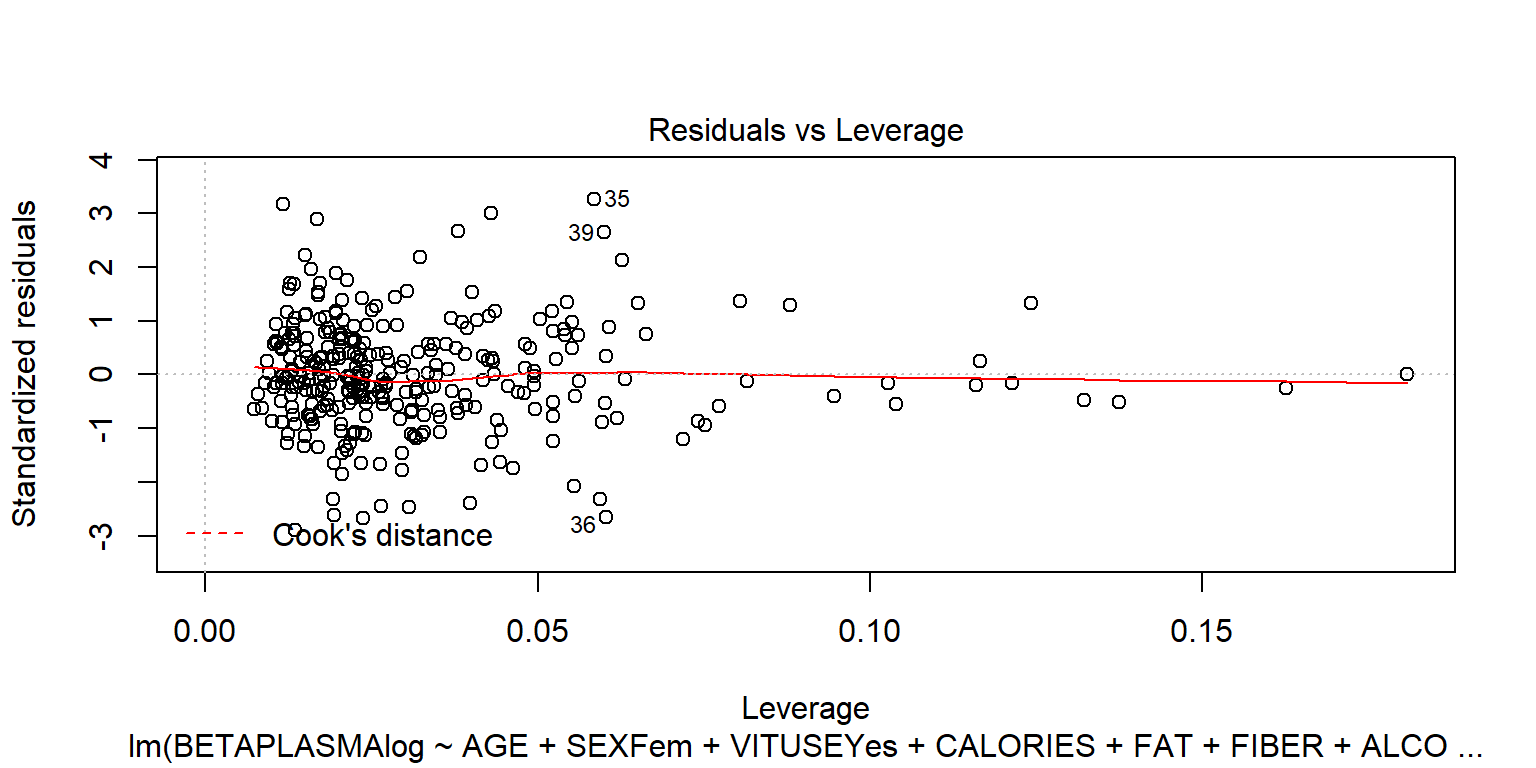
plot(fit2, 4)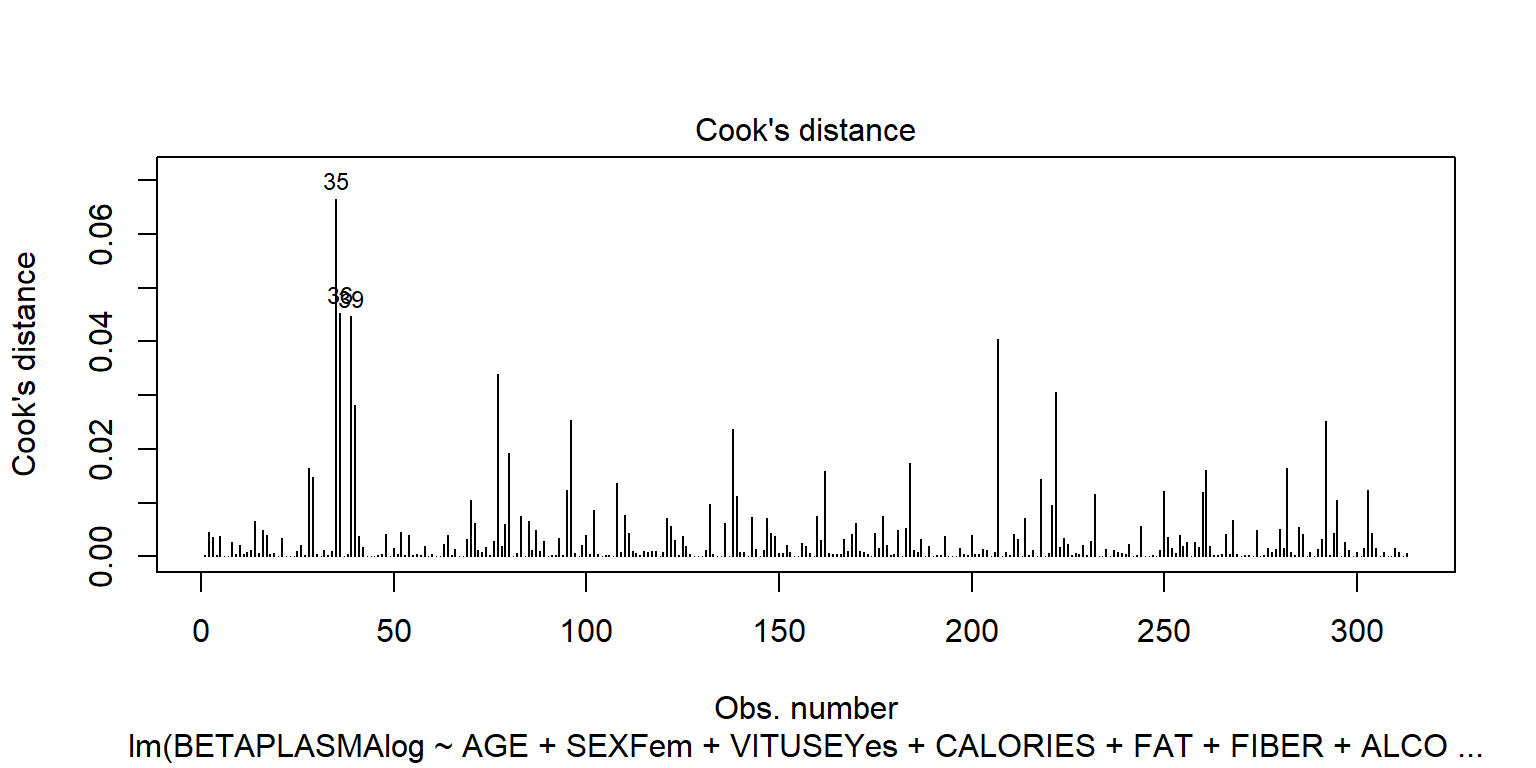
library(broom)
Datosfit2=augment(fit2)
head(Datosfit2)shapiro.test(Datosfit2$.std.resid)##
## Shapiro-Wilk normality test
##
## data: Datosfit2$.std.resid
## W = 1, p-value = 0.003library(nortest)
nortest::lillie.test(Datosfit2$.std.resid)##
## Lilliefors (Kolmogorov-Smirnov) normality test
##
## data: Datosfit2$.std.resid
## D = 0.05, p-value = 0.1library(normtest)
normtest::jb.norm.test(Datosfit2$.std.resid)##
## Jarque-Bera test for normality
##
## data: Datosfit2$.std.resid
## JB = 12, p-value = 0.009```